 |
|||||||||
|
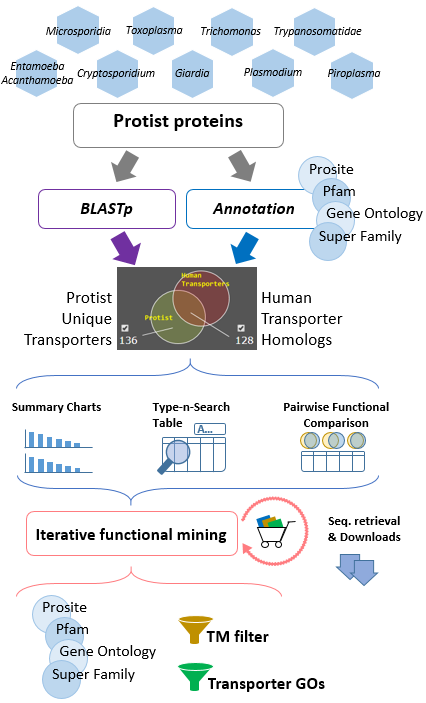
Pathogenic Protist Transmembrane database Pathogenic protist membrane transporter proteins play important roles which notonly on exchanging molecules outside and inside cells but also on inquiring nutrients and biosynthetic compounds from their hosts. Currently, no centralized protist membrane transporter database published makes system-wised comparison and studies of host-pathogen membranome difficult to achieve. We analyzed over one million protein sequences from 139 protists with or partial genome sequenced. Putative transmembrane proteins were annotated by primary sequence alignments, conserved secondary structural elements, and functional domains. Based on these information, a database system with user-friendly interactive data acquiring interfaces implemented by modern front- and back-end cloud-based technologies was built. PPTdb is a web-based database which delivers a user-friendly searching and data querying interface including hierarchical transporter classification number (TC), protein sequences, functional annotations, conserved functional domains, batch sequence retrieving and downloads. PPTdb also serves as an analytic platform which provides useful comparison/mining tools including transmembrane ability evaluation, annotation of unknown proteins, informative visualization charts, and iterative functional mining of host-pathogen transporter proteins. Database statistics
|
||||||||
|
©2018 CGU CSIE Contact
|
|||||||||